RNA-BASED BIOPRODUCTS FOR IMPROVEMENT OF GRAIN YIELD AND QUALITY IN LEGUMES (RIBOLEG)
Bioproductos basados en ARN para el mejoramiento de la producción y calidad de granos de leguminosas (RIBOLEG)
María Eugenia Zanetti, Flavio Blanco, Federico Ariel, Gabriela Michavila, Nadia R. Chalfoun
In the context of the growing demand for quantity and quality of food to meet the needs of a growing population, biotechnology applied to agriculture faces important challenges to develop strategies and bioproducts that allow increasing agricultural productivity in a sustainable manner, without generating harmful impacts on the environment. During the last century, the application of a technological package implemented during the so-called "First Green Revolution" allowed limiting the losses caused by pathogenic microorganisms and overcoming nutrient deficiencies through the application of synthetic pesticides and chemical fertilizers, respectively. This strategy had a strong positive impact on crop productivity, especially in high-income countries; however, for small farmers in low-income countries, the high cost of pesticides and fertilizers created a significant gap in the size of production. On the other hand, the indiscriminate use of synthetic pesticides and chemical fertilizers in agriculture brought about significant harmful effects on the environment. Pesticides can affect the ecosystem immediately, generate adverse effects on human health, or be harmful to the environment in the long term. Chemical nitrogen fertilizers contribute to the production of greenhouse gases, and consequently to global warming. In addition, chemical fertilization is one of the main sources of pollution in agriculture, causing loss of diversity in nearby ecological systems and contaminating water courses such as seas, rivers, and lakes, and directly affecting human health. In this context, modern plant biotechnology faces the great challenge of developing sustainable alternative strategies that allow increasing food production without generating a negative impact on the environment or on the health of agricultural workers, the inhabitants of nearby towns, and of consumers. Technologies based on the use of exogenously applied RNA have emerged as highly promising to solve various aspects of agriculture and improve crop yields without the need to generate elite genetically modified or edited organisms (GMO or GEO, respectively) that replace the great diversity of crop local landraces.
In this project we propose to develop novel and environmentally friendly technologies (clean, innocuous and organic) to, on the one hand, restrict the growth of pathogenic microorganisms, and on the other, optimize nitrogen fixation in four leguminous grain plants of agronomic and regional interest for Argentina (beans, soybeans, chickpeas and peanuts). We understand that the application of these biotechnologies will have a positive and direct impact on the yield of grains and their protein content. These technologies are non-transgenic and are based on the exogenous application of RNAs. By absorbing exogenous double stranded RNAs, these molecules will be processed by the internal machinery of the plant, generating small RNAs that will subsequently silence genes of the plant or of some interacting microorganism (either pathogen or symbiont). This means that the exogenous application of these double-stranded RNAs will restrict the growth of pathogens in crops of agricultural interest, or eventually induce the predisposition of the plant to establish interactions with beneficial microorganisms. Thus, knowing the crop genome, as well as the microorganisms and the human genomes, we can design RNAs and suitable formulations, which could be used in topical applications, as a sustainable alternative, to modulate the microbiome of the crop. Another type of RNAs that can be used as non-transgenic tools to provide information to plants are long noncoding RNAs (that is, they are not messenger RNAs that code for proteins, but perform functions directly as RNA molecules) that modulate in a non-transgenic manner the activity of specific genes. Thanks to its versatility and innocuousness, and its consequent potential in pharmacology and agriculture, RNA is emerging as the central molecule of 21st century biotechnology.
EPIGENETIC AND EPITRANSCRIPTOMIC MARKS AFFECTING RNA TRANSCRIPTION, STABILITY, AND TRANSLATABILITY DURING THE DEVELOPMENT OF LATERAL ROOT ORGANS IN LEGUMES
Marcas epigenéticas y epitranscriptómicas que afectan la transcripción, estabilidad y traducibilidad de RNAs durante el desarrollo de órganos laterales de raíces de leguminosas
María Eugenia Zanetti (PI), Flavio Blanco (CoPI), Mauricio Reynoso (CoPI), Soledad Traubenik (Postdoc), Cristina Kirolinko (PhD student), Milagros Ferrari (PhD student), Jesica Rivero (undergraduate student) Marianela Cueva (undergraduate student), Milagros Yacullo (undergraduate student) and Camila Coronel (undergraduate student)
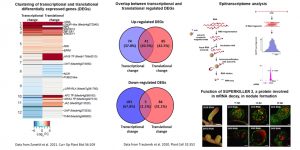
Legume plants form two types of post-embryonic organs in their roots: lateral roots and nodules. Lateral roots serve for soil anchor, water, and nutrient acquisition, whereas nodules are the result of a symbiotic interaction with soil bacteria that allow legumes to overcome nitrogen deficiencies in the soil. Both types of organs have a direct impact on plant growth and development and constitute important agricultural traits. Our research is focused on the characterization of the transcriptional and translational reprograming of root cells during the formation of both types of lateral root organs, as well as changes in mRNA stability mediated by long and small non-coding RNAs during these two developmental processes. Our current aims are to understand the dynamics of epigenetic marks- DNA methylation and histone modifications- that affect chromatin accessibility at transcriptionally active loci during lateral root and nodule organogenesis. In addition, we aim to elucidate how epitranscriptomic marks -particularly N-6 Adenosine methylation (m6A)- affect RNA stability and translatability during the formation of both lateral organs. We use genetic and biochemical approaches combined with high throughput sequencing technologies to elucidate the function of proteins that are crucial for these cellular processes. These results will contribute to a better understanding of the multitier mechanisms that governs the reprogramming of gene expression during the formation of lateral roots and nitrogen fixing nodules. From a biotechnological perspective, this information can be translated into the development of novel strategies for improvement of crop productivity in agriculture.
MULTI-LEVEL REGULATORY MECHANISMS GOVERNING STRAIN SPECIFICITY IN NITROGEN-FIXING SYMBIOSIS
Circuitos regulatorios que controlan la expresión génica en tipos celulares específicos durante la selectividad de cepa en la interacción simbiótica fijadora de nitrógeno
Flavio Blanco (PI), María Eugenia Zanetti (CoPI), Mauricio Reynoso (CoPI), Claudio Rivero (PhD student), Carla Roda (PhD student), Jennifer Artunián (PhD student), Andrés Eylenstein (PhD student), Lourdes Nassarov (undergraduate student) and Marina Cretton (undergraduate student)
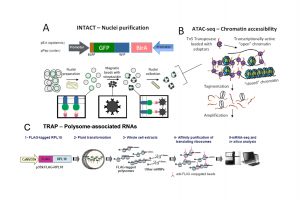
During the legume-rhizobia interaction, root cells undergo a genetic reprogramming that results in the execution of two independent genetic programs that are spatially and temporarily coordinated: infection and the formation of a post-embryonic organ, the nodule, where nitrogen fixation takes place. Biological Nitrogen fixation has been used in agriculture by introducing legumes in crop rotation together with selected strains of rhizobia optimized for nitrogen fixation. To understand genetic changes during this biological process, cell-specific responses, and the multiple layers of gene regulation, from transcriptional to translational level, should be analyzed and integrated. We aim to dissect such responses in epidermal and cortical cells, which are involved in infection and nodule organogenesis, respectively. Genetic reprogramming is evaluated at transcriptional, post-transcriptional and translational levels, including changes in chromatin remodelling, DNA methylation, mRNA dynamics and its translational status. We apply INTACT (A) to purify cell-specific nuclei, ATAC (B), a technique used to characterize dynamic changes in the chromatin state, bisulfite sequencing to assess the dynamics of DNA methylation, RNA-seq, small RNA-seq for transcriptomics and TRAP (C) to study the translational level. Using this information, we have identified regulatory events associated with the efficiency of the symbiotic interaction, but also those that are common across the different accession x strain interactions. Currently, we are functionally characterizing several genes involved in the strain-specific response. These results could help to develop breeding programs that optimize symbiotic outcomes in crop species by improving the compatibility between both symbiotic partners. In the long term, this will contribute to improve agricultural productivity and sustainability by reducing the use of nitrogen fertilizers.
CONSERVED REGULATORY LANDSCAPE OF ROOT LEGUME PLASTICITY
Control conservado de la plasticidad de las raíces de leguminosas
Mauricio Reynoso (PI), María Eugenia Zanetti (CoPI), Flavio Blanco (CoPI), Agustina Tati (undergraduate student) and Camila Bori (undergraduate student)
Increased frequency of extreme climatic events such as floods and droughts affect plant development on both natural and agronomical environments. Plants have the unique capacity of adapting their development to cope with biotic and abiotic stresses derived from these changing conditions. Under this context, the global plant biology community has progressed toward understanding strategies required for plant resilience and aims to increase crop productivity while reducing a negative environmental impact.
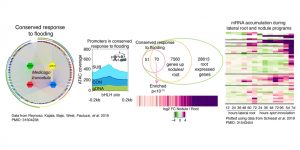
Legumes establish a symbiotic association with rhizobia bacteria capable of fixing atmospheric nitrogen. The interaction leads to the cellular reprogramming of root cells to form post-embryonic organs called nodules, which provide a low oxygen compartment required for nitrogen fixation. Previous studies have found that conserved regulatory networks to flooding across species include a low oxygen signature response. We aim to characterize regulatory mechanisms that could involve transcription factors (TFs) conserved in low oxygen response with focus on their role in cellular reprogramming for lateral root development under floods and nodule development during legume-rhizobia interaction. Evidence supports that most cis-regulatory elements are allocated in chromatin accessible regions present in different cell-populations. We use of a combination of INTACT and ATAC-seq on cells required for lateral root and nodule development that experiment low oxygen conditions to dissect regulatory changes exerted by these TFs. Conserved TFs showing a role in the cellular reprograming are being further evaluated using chromatin immunopurification. This new information will be integrated in the context of already described pathways to provide a better understanding of the key biological processes required for plant root plasticity